UMass College of Natural Sciences Research Greenhouse
Today started out by visiting the growth room on the 12th floor and selecting plants to collect seeds from. We were collecting the T0 seeds from both arabidopsis wildtype and the feronia line. The seed collection process uses newspaper and shaking the seed pods to release the seeds onto the newspaper. Then you use a sifter, which allows the seeds through but keeps most of the debris from passing through. In this way, you are able to get as many seeds as possible while minimizing the debris.
Collected seeds in eppendorf tubes
One of the most important aspects of seed collection is the potential for contamination. You must clean up your work bench, because if seeds from a previous line enter your tube for the new line, it can be catastrophic for results, and can cause a lot of confusion in regards to results.
After we collected seeds, Norice came into lab, and we started running a gel. We used 15 microliters instead of 10, because we were having some slight issues seeing the results earlier in the week. We left the gel running, and analyzed other gels. We were able to tell which of the genes were homozygous and which of the genes were heterozygous. I haven’t read a gel in years, so this was a great review for me.
Gel print-outs; can you tell which plants are homozygous vs heterozygous?
We then spent time in the greenhouse in the afternoon, collecting pollen from the tobacco plants. The process was very straightfoward- remove the tobacco flowers with the most pollen on their anthers and lay them in a tray. I worked collecting the wildtype pollen (wildtype means “normal”) and Norice collected the pollen from the transgenic plants. We went back to the lab with these flowers, and started to empty the pollen into eppendorf tubes.
Tobacco in research greenhouse
Collected tobacco, ready for pollen extraction in the lab.
We then tried to analyze our gel, but there were no bands on the gel. Something had gone wrong in the PCR process, and we were not sure what it was. It ended up being time for me to leave, so I’m not sure what could have gone wrong in our PCR.
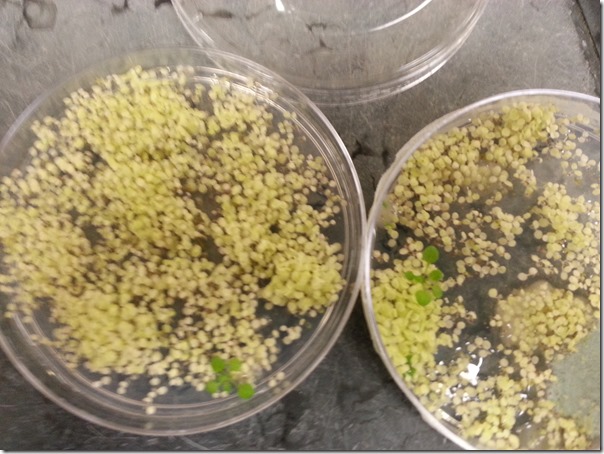
Good example of a selection plate- the green plants are the transformed ones that survived the screening process due to antibiotic resistance. The medium on these plates has antibiotics, which kills the other plants, which are more yellow in this picture.